Posts
Showing posts from July, 2013
Posted by
Llama
Rename error in Linux VirtualLeaf tutorial: "Bareword "tutorial0" not allowed while "strict subs" in use at (eval 1) line 1."
- Get link
- Other Apps
Posted by
Frogee
Nature News article: Quantum boost for artificial intelligence
- Get link
- Other Apps
Posted by
Frogee
Beginnings with Git
- Get link
- Other Apps
Posted by
Frogee
Advice from a systems software engineer
- Get link
- Other Apps
Posted by
Llama
compile errors in Linux VirtualLeaf installation
- Get link
- Other Apps
Posted by
Frogee
Configuring an IGV Genome Server
- Get link
- Other Apps
Posted by
Frogee
On the number of markers and the number of individuals
- Get link
- Other Apps
Posted by
Frogee
Determine size of folders and files in directory
- Get link
- Other Apps
Posted by
Frogee
Extracting sequence information from Stacks samples
- Get link
- Other Apps
Posted by
Frogee
Search and replace in a file without opening it
- Get link
- Other Apps
Posted by
Frogee
Apache error log location in Linux Ubuntu 12.10
- Get link
- Other Apps
Posted by
Frogee
Compression of files in parallel using GNU parallel
- Get link
- Other Apps
Posted by
Frogee
In silico restriction digest and retrieval of restriction site associated DNA
- Get link
- Other Apps
Posted by
Frogee
Determine the type of an object in Python
- Get link
- Other Apps
Posted by
Frogee
Call external program from Python and capture its output
- Get link
- Other Apps
Posted by
Frogee
Installation of Biopieces on Ubuntu Linux 12.10 - Ruby installation
- Get link
- Other Apps
Posted by
Frogee
In silico restriction enzyme digest of eukaryotic genome
- Get link
- Other Apps
Posted by
Frogee
Count number of reads in a SAM file above or below a mapping quality score with awk
- Get link
- Other Apps
Posted by
Frogee
Find CPU information in Ubuntu (12.10)
- Get link
- Other Apps
Posted by
Frogee
Interesting response to Nature Article "Biology must develop its own big data systems"
- Get link
- Other Apps
Posted by
Frogee
Read in file by command line argument and split on tab delimiter in Python
- Get link
- Other Apps
Posted by
Llama
$\LaTeX$ in LibreOffice Impress
- Get link
- Other Apps
Posted by
Frogee
Newbie awk usage example
- Get link
- Other Apps
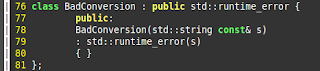
Posted by
Frogee
How to show line numbers in vim
- Get link
- Other Apps

Posted by
Frogee
Write ordered list in LaTeX using enumerate
- Get link
- Other Apps
Posted by
Frogee
How to show LaTex in Blogger
- Get link
- Other Apps
Posted by
Frogee
How to show code snippets in Blogger
- Get link
- Other Apps
Posted by
Llama
smart and graduate studies are mutually exclusive
- Get link
- Other Apps
Posted by
Frogee
Collaborative meeting platform
- Get link
- Other Apps
Posted by
Frogee
Count number of files in directory using bash
- Get link
- Other Apps