Posts
Showing posts from 2016
Posted by
Frogee
Watterson estimator calculation (theta or $\theta_w$) under infinite-sites assumption
- Get link
- Other Apps
Posted by
Frogee
Nucleotide diversity (pi or $\pi$) calculation (per site and per window)
- Get link
- Other Apps

Posted by
Frogee
Postmortem: Chillennium 2016 48-Hour Game Jam - Tardigrade's Dangerous Day
- Get link
- Other Apps

Posted by
Frogee
Postmortem: 3D sorghum reconstructions from depth images identify QTL regulating shoot architecture
- Get link
- Other Apps

Posted by
Frogee
Our first bioRxiv submission
- Get link
- Other Apps
Posted by
Frogee
Robot Hexapod (RHex)
- Get link
- Other Apps

Posted by
Frogee
Getting started with Intel's Threading Building Blocks: installing and compiling an example that uses a std::vector and the Code::Blocks IDE
- Get link
- Other Apps
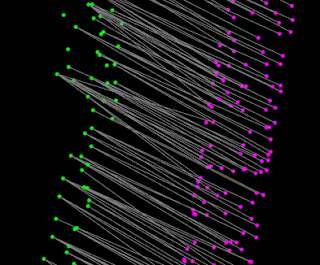
Posted by
Frogee
Access point correspondences from registration method in Point Cloud Library (a.k.a. access protected member in library class without modifying library source).
- Get link
- Other Apps
Posted by
Frogee
Resumes and C.V.s for graduating Ph.D.s
- Get link
- Other Apps