Posts
Showing posts from 2015

Posted by
Frogee
Procedural content generation: map construction via blind agent (a.k.a. building a Rogue-like map)
- Get link
- X
- Other Apps

Posted by
Frogee
Procedural content generation: map construction via binary space partitioning (a.k.a. building a Rogue-like map)
- Get link
- X
- Other Apps
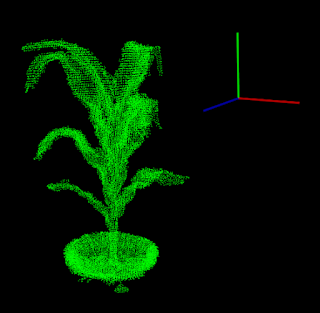
Posted by
Frogee
Rotate 3D vector to same orientation as another 3D vector - Rodrigues' rotation formula
- Get link
- X
- Other Apps
Posted by
Frogee
Desktop Screen Recording and Video Editing (Ubuntu 14.04)
- Get link
- X
- Other Apps

Posted by
Frogee
Find minimum oriented bounding box of point cloud (C++ and PCL)
- Get link
- X
- Other Apps

Posted by
Frogee
Postmortem: RIG: Recalibration and Interrelation of Genomic Sequence Data with the GATK
- Get link
- X
- Other Apps
Posted by
Frogee
Postmortem: Resolution of Genetic Map Expansion Caused by Excess Heterozygosity in Plant Recombinant Inred Populations
- Get link
- X
- Other Apps