Posts
Showing posts from August, 2014
Posted by
Frogee
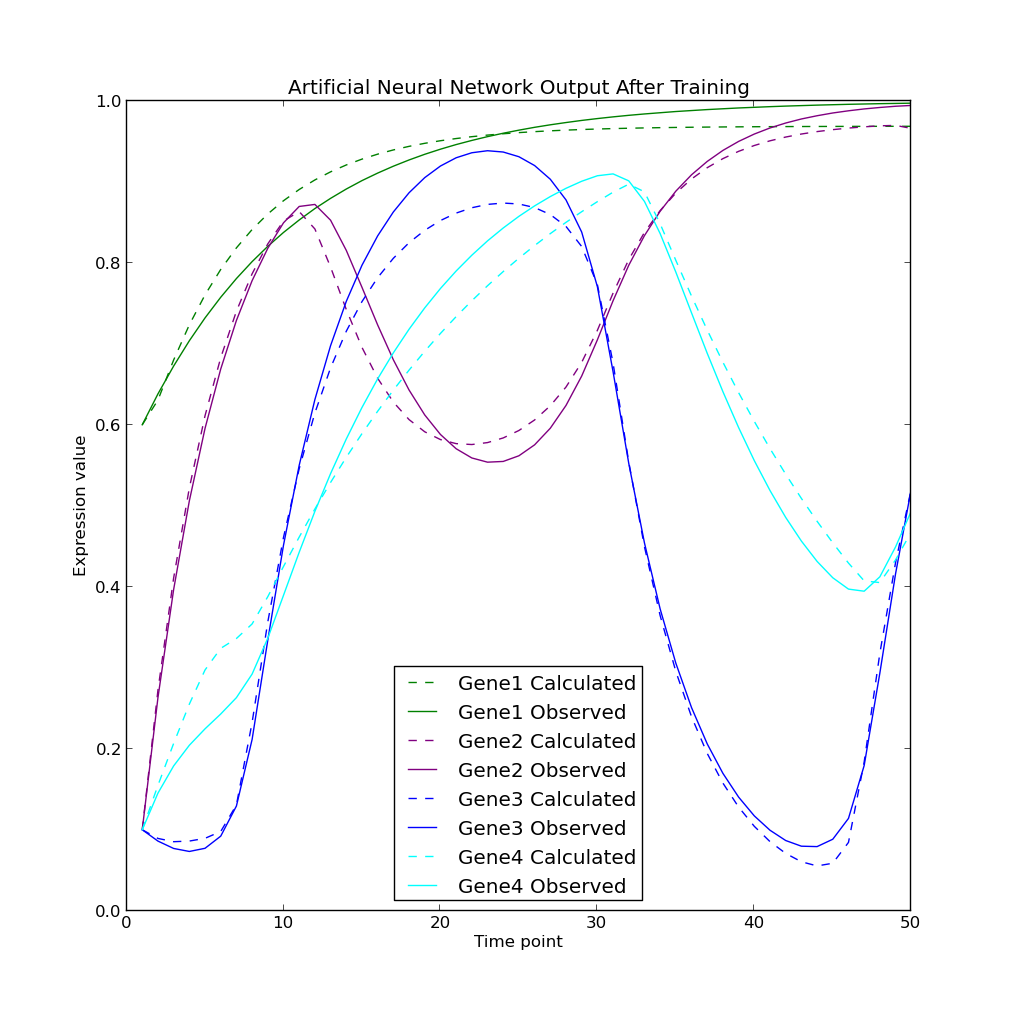
Recurrent artificial neural network evolution using a genetic algorithm for reverse engineering of gene regulatory networks - Postmortem
- Get link
- Other Apps
Posted by
Frogee
Split string on delimiter in C++
- Get link
- Other Apps
Posted by
Frogee
SIAM Conference on the Life Sciences 2014 - Joint Recap and Post Mortem
- Get link
- Other Apps